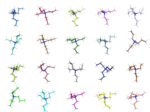2 group members, Elvin and Cameron, have presentations at the virtual ISIMS on twitter this year. Go check out what […]
A brief, condensed tutorial of IMS calculations and simulation is provided in the following jupyter notebook. This example code is […]
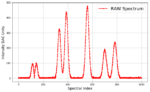
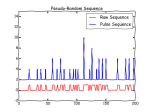
TL/DR — Code and wiring diagram to output a simulated spectrum WITH noise on a specified microcontroller output pin. Requires […]
This is the 3rd post in a series outlining a workflow using freely available computational chemistry resources with python interfaces […]
This is the second post in a series aiming at generating a range of candidate structures for evaluation in the […]
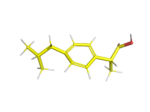
This is the first of a series examining the use of python to generate candidate structures of molecules. These conformations […]
For those that are interested, here is the spreadsheet used in the ASMS 2018 short course. Thank Dr. Bill Siems […]
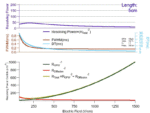
Austen has recently assembled an animation demonstrating the effect of increasing drift tube length on resolving power, calculated from peak […]
For those that are interested, here is the spreadsheet used in the ASMS 2018 short course. Thank Dr. Bill Siems […]
In an effort to create a set of simple tools that are useful for data processing and realtime analysis of […]
Generally, it is recommended to avoid qualitative measures when quantitative solutions exist. However, if the genuine need arises here are […]
Love it or hate it, the lack of a tractable options to create Gantt charts warrants frustration at times. A […]
Now incorporated directly into the latest version of matplotlib (v1.3) here is a great alternative that brings some style to […]